
Dr. Md Mosharof Hosen gained comprehensive exposure to the laboratories across various departments at Sylhet Agricultural University. Through his Master’s degree in Medicine, he developed advanced skills in molecular techniques and laboratory research. Alongside his own thesis work, he actively participated in multiple research projects, which further enhanced his expertise and practical experience in diverse areas of scientific investigation.
Biofilm production and virulence traits among extensively drug-resistant and methicillin-resistant Staphylococcus aureus in buffalo subclinical mastitis in Bangladesh
Role:1st Author (Joint)
Journal: Scientific Reports (Q1, IF:6.5).
Publishing Date: November 2025
Bibliometric Analysis of Research Trends in the Diagnosis of Subclinical Mastitis in Ruminants
Role: Reviewer
Journal: Open Veterinary Journal
Emergence of Multidrug and Extensively Drug-Resistant Escherichia coli and Klebshiella pneumoniae in Subclinical Mastitis of Dairy Buffaloes in Bangladesh: Characterization of Resistance Genes and Biofilm Formation
Role: Co-Author
Conference: 37th BSM Annual Conference, University of Dhaka
Retail Goat Meat as a Source of Multidrug Resistant ESBL-Producing klebsiella pneumoniae : A molecular Investigation
Role: Co-Author
Conference: 37th BSM Annual Conference, University of Dhaka
Epidemiological Insights and Diagnostic Approaches to Neospora caninum Infection in Ruminants: A Comprehensive Review
Role: 1st Author
Conference: Gazipur Agricultural University International Conference 2025, Bangladesh
A farm survey on the prevalence and management practices of Lumpy skin disease in Rajshahi region
Role: Co-Author
Conference: 1st Scientific Conference of Veterinary and Animal Sciences 2025, Rajshahi University
Epidemiology, Modifiable Risk Factors, Therapeutic Managements and Farmer Perceptions of Lumpy Skin Disease in Highly Endemic Regions of Bangladesh
Corresponding Author: Prof. Dr. Md. Mafujur Rahman
Role: Co-author
Journal: Zoonoses and Public Health
Present Status: Submitted
Public Awareness, Perceptions, and Preventive Practices Regarding Mpox in Bangladesh: Insights from a Community-Based Cross-sectional Survey
Corresponding Author: Prof. Dr. Md. Mafujur Rahman
Role: Co-author
Journal: Tropical Medicine & International Health
Present Status: Submitted
Microplastics in Human Fluids, Tissues and Organs: Current Evidence, Detection Technologies, Analytical Challenges, and Health implications.
Corresponding Author: Prof. Dr. Md. Mafujur Rahman
Role: Co-author
Journal: Microplastics
Present Status: Submitted
Cruelty Practices, Animal Welfare, and Hygienic Risks in Live Poultry Retail Systems of Bangladesh. i think it would be extra ordinary
Corresponding Author: Prof. Dr. Md. Mafujur Rahman
Role: Co-author
Present Status: Ongoing
Detection and prevalence of antimicrobial resistance genes in multidrug-resistant and extensively drug-resistant Staphylococcus and Streptococcus species isolated from raw buffalo milk in subclinical mastitis
Responsibilities:
Sample Collection
Laboratory Tests
Isolation, molecular identification and antimicrobial resistance pattern of Methicillin Resistant Staphylococcus aureus (MRSA) isolated from respiratory tract in buffalo.
Responsibilities:
Laboratory Tests
Data Management
Detection of virulence and antimicrobial resistance in E. coli isolated from Buffalo
Responsibilities:
Laboratory Tests
Data Management
Occurrence and Treatment Pattern of common Bacterial Diseases in Pet Animals Practices: Observations form Central Veterinary Hospital, Dhaka
Supervisor: Dr. Md. Rashedunnabi Akanda
• Bacteria culture and isolation
• DNA/RNA extraction (Kit & Boiling method)
• Conventional PCR, Gel electrophoresis
• AST, HA, HI, ELISA
• Vaccine and drug design
• Phylogenetic analysis
• Molecular docking
• Primer design
• Mendeley & Endnote
• MS Office, SPSS, ArcGIS
• Adobe Illustrator, Photoshop, Canva
• PyRx, Bioedit, Pymol, MEGA, ChimeraX
Genomic detection and antibiotic resistance patterns of methicillin-resistant Staphylococcus aureus (MRSA) isolated from subclinical mastitis-affected buffalo milk in Bangladesh
Supervisor: Prof. Dr. Md. Mafujur Rahman
Lab: Medicine Lab, Sylhet Agricultural University
For the detection of Methicillin-Resistant Staphylococcus aureus (MRSA), Dr. Md. Mosharof Hosen collected milk samples from buffaloes across various buffalo-rearing regions in Bangladesh. Samples were transported to the Department of Medicine Laboratory at Sylhet Agricultural University for detailed microbiological and molecular analysis.
Throughout the entire process, he strictly adhered to aseptic techniques to prevent contamination and ensure the reliability of the research results.


Dr. Hosen ensured that all samples were promptly transferred to the Medicine Laboratory of Sylhet Agricultural University immediately after collection from the research areas. To enhance the growth and isolation of microorganisms from the samples, he prepared various types of culture media. The media frequently used in this study included Nutrient Broth, MacConkey Agar, Eosin Methylene Blue (EMB) Agar, Mannitol Salt Agar, and Soybean Casein Digest Medium.
These selective and differential media were essential for enriching and identifying Staphylococcus aureus and other bacteria relevant to the study.
Following media preparation, Dr. Hosen cultured the samples on Petri dishes to facilitate the growth and multiplication of microorganisms. Through this process of culture, subculture, and ultimately obtaining pure cultures, the targeted organisms were isolated. Identification of the organisms was based on colony morphology, pigment production, and a series of biochemical tests.
This systematic approach ensured accurate recognition and characterization of the bacterial species present in the samples.


Identified the bacterial isolates by performing various biochemical tests to examine their characteristic traits. Among the tests routinely conducted were motility, indole and urease tests. Additionally, citrate utilization was evaluated using Simmons’ Citrate Agar and Voges-Proskauer tests were carried out in his laboratory.
These biochemical assessments were crucial for the accurate identification and differentiation of the microorganisms involved in the study.
Dr. Hosen utilized the polymerase chain reaction (PCR) technique to amplify specific DNA sequences of the target bacteria, enabling precise identification. After isolating the organisms, molecular confirmation was performed through PCR analysis.
He detected key virulence genes (hla, icaA, sea, and fnbA) as well as antimicrobial resistance genes (tetA, strA, aac-3(iv), and sul1) by conducting both uniplex and multiplex PCR assays.
This molecular approach provided definitive confirmation of the bacterial isolates and their genetic determinants related to virulence and antibiotic resistance.


Following PCR amplification, the PCR products to gel electrophoresis to separate and analyze the DNA fragments. The gel was then placed under a UV trans-illuminator, allowing the visualization of PCR bands.
The results were documented and analyzed using a computer imaging system, facilitating accurate interpretation of the molecular data.
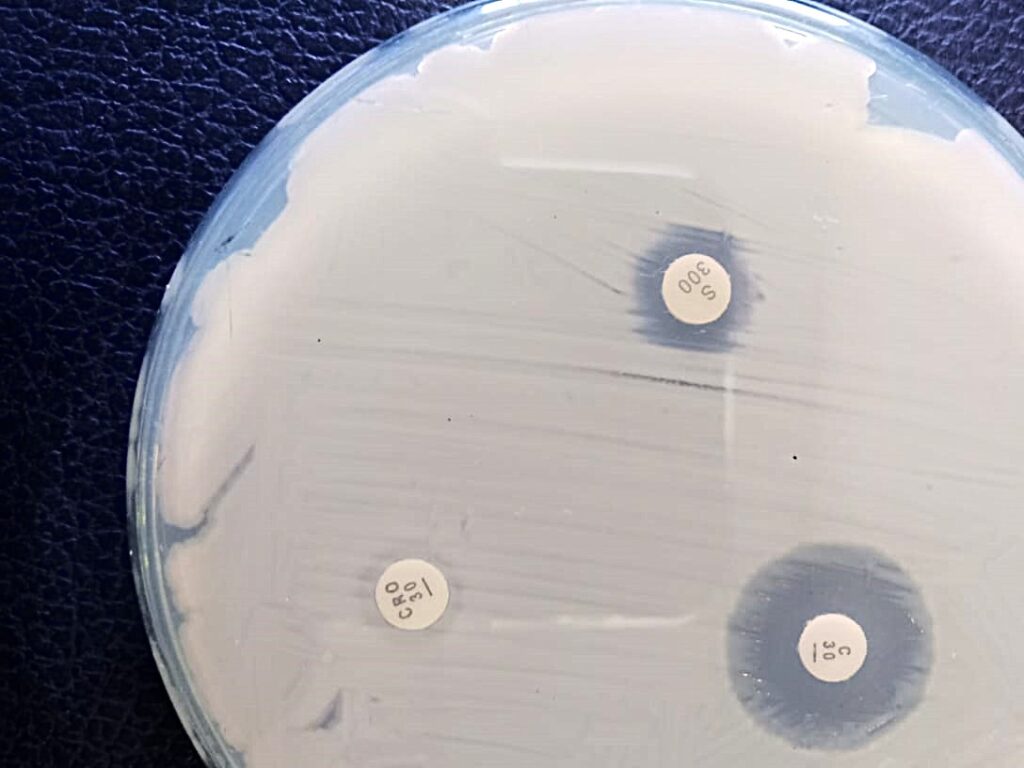
The antibiotic sensitivity of each PCR-positive bacterial isolate was assessed using the Kirby-Bauer disk diffusion method. Twelve commonly used antibiotics were tested to determine the susceptibility patterns of the isolates.
This method provided crucial information on antimicrobial resistance profiles to guide appropriate treatment options.
A dedicated veterinarian and young social activist passionate about serving underprivileged communities.
© 2025 Dr. Md. Mosharof Hosen. All rights reserved.